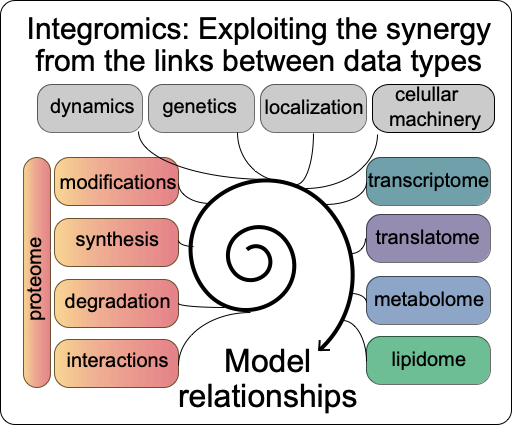
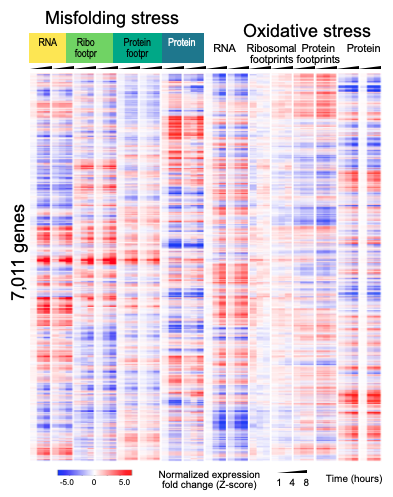
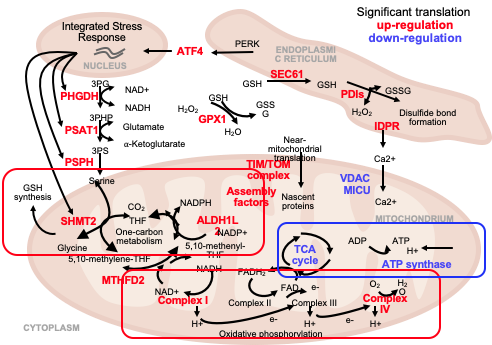
Our main goal is to understand how the synthesis and degradation of mRNAs and proteins are coordinated when cells respond to the accumulation of misfolded proteins. To do so, we employ both experimental and computational approaches, we investigate whole genomes and individual genes, and we probe proteins as well as RNAs.
We have developed the tools to systematically explore the dynamic relationships between sequencing, ribosome footprinting, and proteomics data, and experimental methods to parse small protein samples from as few as 1,000 cells. We have quantified and compared transcription, translation, and protein stability of cells responding to protein misfolding stress. We have discovered new links between small molecule metabolism and the stress response, roles of alternative splicing, and translation regulatory elements. We have demonstrated new roles for the proteasome in induced motor neurons that might help understand Amyotrophic Lateral Sclerosis, a fatal neurodegenerative disease. We continue to investigate different aspects of this system, e.g. translation regulatory elements that affect the cellular stress response in these cells.
A summary of our proteomics research can be found in this talk as part of the Proteostasis Consortium Seminar Series.